Training a Convolutional Neural Network (CNN) for performing medical images/ imaging tasks has always been done using Transfer learning as any other natural image classification task (cats Vs dogs). In other words, to build a medical image classification artificial intelligence model (disease detection, cancer malignancy identification, chest x-ray anomaly detection) we still fine-tune a pre-trained model such as InceptionV3 network, which was trained originally on ImageNet, on another medical dataset. So the issue here is we are fine-tuning a model, for performing a medical image classification task, that was originally trained on natural images like ImageNet. But, have you asked yourselves what can happen if we train the original model on medical images dataset similar to ImageNet, and then we fine-tune it on a specific dataset when needed (also medical)!. RadImageNet
The answer to the question above is that pretraining and fine-tuning using medical images instead of ImageNet can or cannot be an advantage to the medical imaging dataset as it will be trained and fine-tuned on the same image modality. To prove this, a new Open Radiologic Deep Learning Research Dataset for Effective Transfer Learning was proposed to show the advantages of employing transfer learning and pretraining using millions of radiologic/medical images instead of ImageNet natural images for use in downstream medical applications.
The RadImageNet database contains 1.35 million annotated medical images from 131 872 patients who underwent CT, MRI, and US for pathologic conditions affecting the musculoskeletal system, the nervous system, cancer, the gastrointestinal tract, the endocrine system, the abdominal cavity, and the lungs. The RadImageNet models showed a significant advantage (P .001) over ImageNet models for transfer learning tasks on small datasets, including thyroid nodules (US), breast masses (US), anterior cruciate ligament injuries (MRI), and meniscal tears (MRI). The improvements in AUC were 9.4%, 4.0%, 4.8%, and 4.5%, respectively. The RadImageNet models also showed increased AUC (P .001) for bigger datasets, including cerebral hemorrhage (CT), COVID-19 (chest radiography), SARS-CoV-2 (CT), and pneumonia (chest radiography). Additionally, on thyroid and breast US datasets, the RadImageNet models' lesion localizations were enhanced by 64.6% and 16.4%, respectively.
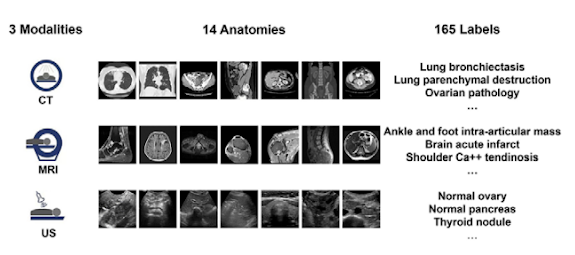
Sample of the RadImageNet images, modalities, and labels
Study and Experiment of RadImageNet
The creators of this dataset showed in their original article that this big dataset can be used for training models from scratch and for fine-tuning pre-trained models. Experimentally, the authors provided a comparison of RadImageNet and ImageNet Pretrained
Models in terms of performance. this comparison aims to answer the question of whether a model pre-trained originally using RadImageNet can perform better than the one pre-trained using ImageNet if fine-tuned using a medical dataset?
In the comparison experiment, the authors used RadImageNet and ImageNet pre-trained models for transfer learning on eight external downstream different applications. For transfer learning, we used RadImageNet and ImageNet pre-trained models on eight downstream external applications.
The following was among the eight tasks: classification of benign and malignant thyroid nodules using ultrasound with 288 malignant and 61 benign images (23); classification of benign and malignant breast lesions using ultrasound with 210 malignant and 570 benign images (24); detection of meniscal and anterior cruciate ligament (ACL) tears using MRI with 570 ACL tear images, 452 non-ACL tear images, 506 meniscal tear images, and 3695 nonmeniscal tear images (25); Chest radiographs were used to detect pneumonia, with 6012 pneumonia chest radiographs and 20 672 nonpneumonia images (26); to distinguish patients with COVID-19 from those with community-acquired pneumonia; with 21872 COVID-19 images and 36894 community-acquired pneumonia images (27); to classify patients with and without COVID-19 at chest CT; with 4190 COVID-19-positive and 4860 COVID-19-negative images (15); and to (28).
Training the Models
Receiver operating curve analysis was used to assess the RadImageNet and ImageNet models. For each application, the models were adjusted using simulations of a total of 24 scenarios. The learning rates and the number of freeze layers used to train the four CNNs vary. While the freezing of all layers and the unfreezing of the top 10 layers were carried out with learning rates of 0.01 and 0.001, respectively, and 0.001 and 0.0001, respectively. RadImageNet and ImageNet pre-trained models were compared based on the average AUC and SD of these 24 values. Each dataset for a downstream application was divided into three parts: a training set, a validation set, and a test set.
The same collection of images was always used for one patient. The loss function chosen was binary cross-entropy. The input photos were reduced in size to 256 3 256 pixels in order to balance accuracy and productivity. After the final layer of the pre-trained models, a global average pooling layer, a dropout layer, and an output layer activated by the softmax function were added. Models underwent 30 epochs of training.
Results and Discussion
1.35 million annotated CT, MRI, and US images of musculoskeletal, neurologic, oncologic, gastrointestinal, endocrine, and pulmonary pathologic abnormalities are included in the RadImageNet dataset. they gathered the most prevalent modalities and anatomical locations on the same scale for direct comparison with ImageNet (the starting size for the ImageNet challenge was 1.3 million photos). Three radiologic modalities, eleven anatomical locations, and 165 pathologic labels make up the RadImageNet database, which was utilized to compare ImageNet to other databases. The results on the Thyroid dataset, as shown by the paper, are shown below:
The results on the Chest COVID-19 dataset, as shown by the paper, are shown below:
Overall, what we can learn from this is that regardless of the sample size of the applications, the RadImageNet models showed superior performance in imaging identification and consistency over 24 simulated tuning scenarios. When compared to unfreezing partial layers and training only completely connected layers within the 24 scenarios, unfreezing all layers consistently produced the greatest performance. When training all trainable parameters, a lower learning rate of 0.0001 would be advised to potentially more accurately capture global optimal performance. ResNet50, DenseNet121, and Inception-ResNet-v2 all outperformed Inception V3 in terms of performance when computational power and turnaround time were available. On tiny datasets, however, RadImageNet models' advantage becomes more obvious.
More about the RadImageNet Dataset: Modalities, Anatomic Regions, and Classes
The RadImageNet database used for comparison to ImageNet consists of three radiologic modalities, eleven
anatomic regions, and 165 pathologic labels as shown the Figures below:
Dataset availability
The RadImageNet pre-trained models, dataset, and codes can be found at https://github.com/BMEII-AI/RadImageNet.







Comments
Post a Comment