Review of Latest Works:
Understanding the Problem:
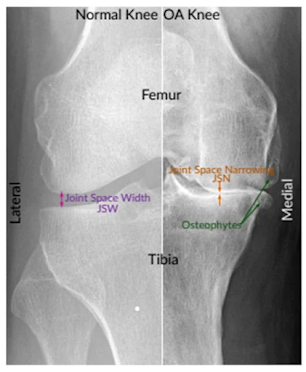
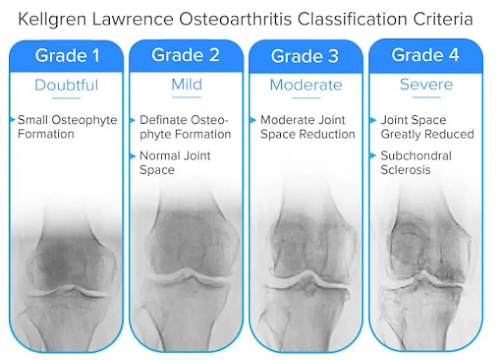
Challenges in Knee OA Grading:
- Limited Labeled Data: Building robust models requires large, diverse, and accurately labeled datasets, which are often scarce in the medical domain.
- Inter-Observer Variability: Different clinicians may interpret the same radiographic images differently, leading to inconsistencies in grading.
- Non-Linearity in Disease Progression: Knee OA is a complex, nonlinear condition with variations in disease manifestation, making it challenging to capture using traditional linear models.
- Interpretability and Explainability: Understanding the decisions of AI models is crucial in medical applications. Ensuring that the model's predictions align with clinical reasoning is essential for widespread adoption.
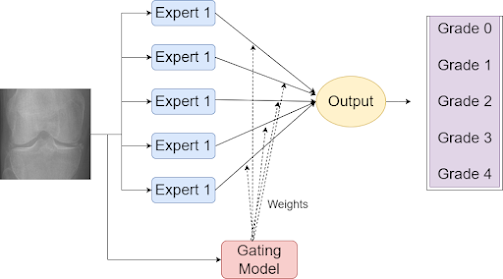
Proposed Solution: Mixture of Experts
Mixture of Experts (MoE) is a neural network architecture that combines the strengths of both global and local models. It consists of expert networks responsible for specific regions of the input space and a gating network that determines the contribution of each expert to the final prediction. This approach enables capturing the intricate patterns present in knee OA images.
Implementation Steps:
- Data Preprocessing:
- Load and preprocess knee OA radiographic images.
- Split the dataset into training, validation, and test sets.
- Model Architecture:
- Design the MoE architecture using TensorFlow or PyTorch.
- Define expert networks and a gating network.
- Train the model on the training dataset.
- Training Strategy:
- Employ techniques like data augmentation to enhance model generalization.
- Implement transfer learning if pre-trained models are available.
- Evaluation:
- Assess the model on the validation set and fine-tune hyperparameters.
- Evaluate the final model on the test set to measure its performance.
- Interpretability:
- Implement methods to interpret and explain the decisions made by the MoE model.
Implementation of Mixture of Experts for Knee Osteoarthritis Grading in TensorFlow
import os
import cv2
import numpy as np
from sklearn.model_selection import train_test_split
from keras.utils import to_categorical
# Function to load and preprocess knee OA radiographic images
def load_and_preprocess_data(base_dir, image_size=(64, 64)):
data = []
labels = []
# Iterate through subdirectories (each subdirectory corresponds to a class)
for class_label in os.listdir(base_dir):
class_path = os.path.join(base_dir, class_label)
for image_file in os.listdir(class_path):
image_path = os.path.join(class_path, image_file)
# Read and resize the image
image = cv2.imread(image_path)
image = cv2.resize(image, image_size)
# Normalize pixel values to be between 0 and 1
image = image.astype('float32') / 255.0
data.append(image)
labels.append(int(class_label)) # Assuming subdirectory names represent class labels
return np.array(data), np.array(labels)
# Load and preprocess knee OA radiographic images
base_dir = r'C:\Users\abdul\Desktop\Research\Knee Osteo\56rmx5bjcr-1\KneeXrayData\KneeXrayData\ClsKLData\kneeKL224'
data, labels = load_and_preprocess_data(base_dir)
# Split the dataset into training, validation, and test sets
X_train, X_temp, y_train, y_temp = train_test_split(data, labels, test_size=0.3, random_state=42)
X_val, X_test, y_val, y_test = train_test_split(X_temp, y_temp, test_size=0.5, random_state=42)
# Convert labels to one-hot encoding
num_classes = 5 # Assuming 5 classes for KOA grading
y_train_one_hot = to_categorical(y_train, num_classes)
y_val_one_hot = to_categorical(y_val, num_classes)
y_test_one_hot = to_categorical(y_test, num_classes)# Expert Model
def create_expert_model(input_shape, num_outputs):
model = models.Sequential([
layers.Conv2D(32, (3, 3), activation='relu', input_shape=input_shape),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(64, (3, 3), activation='relu'),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(128, (3, 3), activation='relu'),
layers.MaxPooling2D((2, 2)),
layers.Flatten(),
layers.Dense(128, activation='relu'),
layers.Dense(num_outputs, activation='softmax')
])
return model# Gating Model
def create_gating_model(num_experts, num_outputs):
model = models.Sequential([
layers.Dense(128, activation='relu', input_shape=(num_experts,)),
layers.Dense(num_outputs, activation='softmax')
])
return model# Mixture of Experts (MOE) Model
def create_moe_model(input_shape, num_experts, num_outputs):
expert_input = layers.Input(shape=input_shape, name='expert_input')
expert_model = create_expert_model(input_shape, num_outputs)
expert_output = expert_model(expert_input)
gating_input = layers.Input(shape=(num_experts,), name='gating_input')
gating_model = create_gating_model(num_experts, num_outputs)
gating_output = gating_model(gating_input)
# Reshape gating output to match expert output shape
gating_output = layers.Reshape((num_outputs, 1))(gating_output)
gating_output = layers.Lambda(lambda x: layers.Flatten()(x))(gating_output)
# Multiply expert and gating outputs
mixture_output = layers.Multiply()([expert_output, gating_output])
moe_model = models.Model(inputs=[expert_input, gating_input], outputs=mixture_output)
return moe_modelTraining Strategy:
# Assuming you have already defined the create_moe_model function from the previous response
# Specify input shape, number of experts, and number of classes
input_shape = (64, 64, 1) # Input image dimensions
num_experts = 5 # number of experts
num_classes = 5 # Number of KOA grading classes
# Create the Mixture of Experts (MOE) model
moe_model = create_moe_model(input_shape, num_experts, num_classes)
# Compile the model
moe_model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
# Train the model
epochs = 20
batch_size = 32
history = moe_model.fit(
[X_train, np.random.rand(len(X_train), num_experts)],
y_train_one_hot,
epochs=epochs,
batch_size=batch_size,
validation_data=([X_val, np.random.rand(len(X_val), num_experts)], y_val_one_hot)
)
# Evaluate the model on the test set
test_loss, test_accuracy = moe_model.evaluate([X_test, np.random.rand(len(X_test), num_experts)], y_test_one_hot)
print(f'Test Accuracy: {test_accuracy * 100:.2f}%')Loss Function:
- X_train: This is our actual input data, presumably the knee osteoarthritis radiographic images.
- np.random.rand(len(X_train), num_experts): This is a random array generated using NumPy. It creates an array with a shape of (len(X_train), num_experts). This array is used as additional input, typically for the gating network in the mixture-of-experts model.
Training and Evaluation
Epoch 1/20
157/157 [==============================] - 12s 73ms/step - loss: 1.4890 - accuracy: 0.3559 - val_loss: 1.4881 - val_accuracy: 0.3684
Epoch 2/20
157/157 [==============================] - 11s 70ms/step - loss: 1.4773 - accuracy: 0.3591 - val_loss: 1.4840 - val_accuracy: 0.3684
Epoch 3/20
157/157 [==============================] - 8s 50ms/step - loss: 1.4614 - accuracy: 0.3608 - val_loss: 1.4230 - val_accuracy: 0.3721
Epoch 4/20
157/157 [==============================] - 11s 70ms/step - loss: 1.3706 - accuracy: 0.3838 - val_loss: 1.3056 - val_accuracy: 0.4260
Epoch 5/20
157/157 [==============================] - 9s 57ms/step - loss: 1.2741 - accuracy: 0.4222 - val_loss: 1.2533 - val_accuracy: 0.4567
Epoch 6/20
157/157 [==============================] - 10s 64ms/step - loss: 1.2253 - accuracy: 0.4492 - val_loss: 1.2030 - val_accuracy: 0.4679
Epoch 7/20
157/157 [==============================] - 11s 70ms/step - loss: 1.1856 - accuracy: 0.4661 - val_loss: 1.2973 - val_accuracy: 0.3777
Epoch 8/20
157/157 [==============================] - 9s 57ms/step - loss: 1.1566 - accuracy: 0.4821 - val_loss: 1.1579 - val_accuracy: 0.4930
Epoch 9/20
157/157 [==============================] - 12s 74ms/step - loss: 1.1304 - accuracy: 0.4880 - val_loss: 1.1321 - val_accuracy: 0.4940
Epoch 10/20
157/157 [==============================] - 11s 71ms/step - loss: 1.1121 - accuracy: 0.5040 - val_loss: 1.1053 - val_accuracy: 0.5107
Epoch 11/20
157/157 [==============================] - 11s 69ms/step - loss: 1.0854 - accuracy: 0.5163 - val_loss: 1.1183 - val_accuracy: 0.5033
Epoch 12/20
157/157 [==============================] - 11s 71ms/step - loss: 1.0756 - accuracy: 0.5148 - val_loss: 1.1826 - val_accuracy: 0.4735
Epoch 13/20
157/157 [==============================] - 12s 75ms/step - loss: 1.0611 - accuracy: 0.5249 - val_loss: 1.0969 - val_accuracy: 0.5107
Epoch 14/20
157/157 [==============================] - 8s 52ms/step - loss: 1.0345 - accuracy: 0.5373 - val_loss: 1.1147 - val_accuracy: 0.4995
Epoch 15/20
157/157 [==============================] - 11s 67ms/step - loss: 1.0219 - accuracy: 0.5441 - val_loss: 1.1329 - val_accuracy: 0.5228
Epoch 16/20
157/157 [==============================] - 10s 61ms/step - loss: 1.0286 - accuracy: 0.5419 - val_loss: 1.1324 - val_accuracy: 0.4223
Epoch 17/20
157/157 [==============================] - 9s 60ms/step - loss: 1.0015 - accuracy: 0.5486 - val_loss: 1.0764 - val_accuracy: 0.5144
Epoch 18/20
157/157 [==============================] - 13s 82ms/step - loss: 0.9814 - accuracy: 0.5590 - val_loss: 1.0627 - val_accuracy: 0.5293
Epoch 19/20
157/157 [==============================] - 10s 61ms/step - loss: 0.9768 - accuracy: 0.5702 - val_loss: 1.1044 - val_accuracy: 0.5126
Epoch 20/20
157/157 [==============================] - 8s 52ms/step - loss: 0.9527 - accuracy: 0.5712 - val_loss: 1.0604 - val_accuracy: 0.5200
34/34 [==============================] - 1s 15ms/step - loss: 1.0263 - accuracy: 0.5428
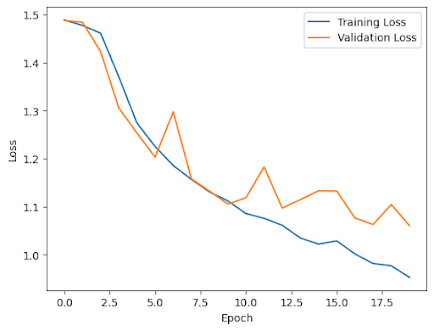
Test Accuracy: 54.28%import matplotlib.pyplot as plt
plt.plot(history.history['loss'], label='Training Loss')
plt.plot(history.history['val_loss'], label='Validation Loss')
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.legend()
plt.show()import tensorflow as tf
import numpy as np
import cv2
from matplotlib import pyplot as plt
def preprocess_image(img_path):
img = tf.keras.preprocessing.image.load_img(img_path, target_size=(224, 224))
img_array = tf.keras.preprocessing.image.img_to_array(img)
img_array = np.expand_dims(img_array, axis=0)
img_array = tf.keras.applications.mobilenet_v2.preprocess_input(img_array)
return img_array
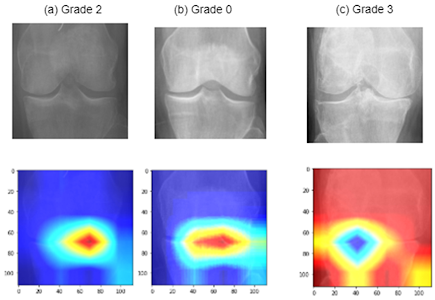
def get_grad_cam(model, img_array, layer_name):
grad_model = tf.keras.models.Model(
[model.inputs], [model.get_layer(layer_name).output, model.output]
)
with tf.GradientTape() as tape:
conv_outputs, predictions = grad_model(img_array)
loss = predictions[:, np.argmax(predictions[0])]
grads = tape.gradient(loss, conv_outputs)[0]
guided_grads = tf.cast(conv_outputs > 0, 'float32') * tf.cast(grads > 0, 'float32') * grads
weights = tf.reduce_mean(guided_grads, axis=(0, 1))
cam = tf.reduce_sum(tf.multiply(weights, conv_outputs), axis=-1)
return cam.numpy()
def overlay_grad_cam(img_path, cam, alpha=0.4):
img = cv2.imread(img_path)
img = cv2.resize(img, (224, 224))
heatmap = cv2.resize(cam, (img.shape[1], img.shape[0]))
heatmap = np.uint8(255 * heatmap)
heatmap = cv2.applyColorMap(heatmap, cv2.COLORMAP_JET)
superimposed_img = cv2.addWeighted(img, alpha, heatmap, 1 - alpha, 0)
return superimposed_img
# Load the trained model
model = tf.keras.models.load_model('your_model_path.h5')
# Load and preprocess the image
img_path = 'image1.jpg'
img_array = preprocess_image(img_path)
# Choose the layer for which you want to visualize the activation map
layer_name = 'Conv1'
# Get the Grad-CAM
cam = get_grad_cam(model, img_array, layer_name)
# Overlay Grad-CAM on the original image and display
result = overlay_grad_cam(img_path, cam)
plt.imshow(result)
plt.axis('off')
plt.show()
Comments
Post a Comment